Multiwfn forum
Multiwfn official website: //www.umsyar.com/multiwfn. Multiwfn forum in Chinese: http://bbs.keinsci.com/wfn
You are not logged in.
- Topics:Active|Unanswered
#12022-06-24 11:26:41
- sahop
- Member
- Registered: 2021-08-28
- Posts: 3
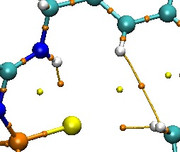
No bond paths appear around the sulfur atom in QTAIM diagram
Dear Tian Lu
I tried to plot the QTAIM diagram between some molecules using MultiWFN. To do this analysis, I used the .molden files generated by the NWCHEM software and then converted them to WFN files. In the final results, everything is ok, but only the bond paths do not appear around the sulfur atom. (An image of the problem is attached.)
I have done this analysis with other molecules before (Whether with the NWCHEM output or with other software outputs, but I have not had this problem.
I would be grateful if you could guide me.
best regards
Saeed Hosseinpoor
Offline
#22022-06-24 12:22:15
Re: No bond paths appear around the sulfur atom in QTAIM diagram
Dear Saeed Hosseinpoor,
Please first check if (3,-3) CP at nuclear position of the sulfur has been found. If it was not found, then bond path cannot be generated.
Also, please show me the basis set you used in your NWChem calculation.
Note that Multiwfn directly supports .molden file generated by NWChem, this point is mentioned in beginning of Chapter 4 of Multiwfn (see below), converting it to .wfn format is redundant.
NWChem: An example input file is provided as examples\NWChem_molden.nw. After running it, the .molden file will be generated in current folder. Notice that spherical harmonic basis functions must be employed (i.e. the "spherical" keyword) and "noautosym" keyword must be employed when the system has symmetry of point group.
Best regards,
Tian
Offline
#32022-06-24 16:21:24
- sahop
- Member
- Registered: 2021-08-28
- Posts: 3
Re: No bond paths appear around the sulfur atom in QTAIM diagram
Dear Tian
The sulfur (3, -3) CPs exist.
I use 6-31G** basis set for my calculations.
Offline
#52022-06-25 13:56:28
Re: No bond paths appear around the sulfur atom in QTAIM diagram
I have checked your file, your molden file is problematic. You can use the way mentioned in Appendix 5 of Multiwfn manual to check sanity of the file, you will find the wavefunction loaded into Multiwfn is incorrect.
As I mentioned in 2#, please add spherical and noautosym keywords and generate the molden file again, and then test if the new molden file can pass check.
Offline