Multiwfn forum
Multiwfn official website: //www.umsyar.com/multiwfn. Multiwfn forum in Chinese: http://bbs.keinsci.com/wfn
You are not logged in.
- Topics:Active|Unanswered
#12023-06-18 16:12:57
- amin.alibakhshi@hotmail.c
- Member
- Registered: 2019-08-25
- Posts: 23
some potential bugs
Dear Prof. Lu, Dear developers of Multiwfn,
I just recently noticed two issues that seems to me like a bug.
In the calculation of Fuzzy atomic space analysis based on the Hirshfeld-I method, for cases Gaussian is needed to compute .rad files, while g09 works perfectly, it doesn't work for the g16 version and I suppose the problem stems from the way .gjf is written.
The second issue is related to the same computations when H atom is involved. For that, some errors are generated for H+1 computations which is kind of natural as there is no electron and therefore, wfn or .rad file is not defined in theory.
Kind regards + thank you for your wonderful job
Last edited by amin.alibakhshi@hotmail.c (2023-06-18 16:17:24)
Offline
#22023-06-19 03:04:24
Re: some potential bugs
Hello,
How did you calculate the.rad files? If you directly use Multiwfn to automatically invoke Gaussian 16 to calculate the .rad files, I don't find problem, see relevant output of my test:
============== Generate Hirshfeld-I atomic weights ============== -3 Switch if speeding up calculation using distance cutoff, current: Yes, ratio factor is 2.000 -2 Switch algorithm, current: Fast & large memory requirement 1 Start calculation! 2 Set the maximum number of iterations, current: 50 3 Set convergence criterion of atomic charges, current: 0.000200 1 Some atomic .wfn files are not found in "atmrad" folder in current directory Now input the level for calculating these .wfn files, e.g. B3LYP/def2SVP You can also add other keywords at the same time, e.g. M062X/6-311G(2df,2p) scf=xqc int=ultrafine b3lyp/6-31G* Running: "/sob/g16/g16" "atmrad/C-2.gjf" "atmrad/C-2" Running: "/sob/g16/g16" "atmrad/C-1.gjf" "atmrad/C-1" Running: "/sob/g16/g16" "atmrad/C_0.gjf" "atmrad/C_0" Running: "/sob/g16/g16" "atmrad/C+1.gjf" "atmrad/C+1" Running: "/sob/g16/g16" "atmrad/C+2.gjf" "atmrad/C+2" Running: "/sob/g16/g16" "atmrad/H-1.gjf" "atmrad/H-1" Running: "/sob/g16/g16" "atmrad/H_0.gjf" "atmrad/H_0" Running: "/sob/g16/g16" "atmrad/H+1.gjf" "atmrad/H+1" Converting atmrad/C-2.wfn to atmrad/C-2.rad Converting atmrad/C-1.wfn to atmrad/C-1.rad Converting atmrad/C_0.wfn to atmrad/C_0.rad Converting atmrad/C+1.wfn to atmrad/C+1.rad Converting atmrad/C+2.wfn to atmrad/C+2.rad Converting atmrad/H-1.wfn to atmrad/H-1.rad Converting atmrad/H_0.wfn to atmrad/H_0.rad Converting atmrad/H+1.wfn to atmrad/H+1.rad Radial grids: 75 Angular grids: 434 Total: 32550 After pruning: 26474 Calculating actual density of system at all grids... Progress: [##################################################] 100.0 % - Memory requirement for storing atomic densities on grids: 43.5 MB Calculating atomic density contribution to grids... Progress: [##################################################] 100.0 % \.rad H+1 state doesn't have an electron, if you open the .rad file generated by Multiwfn, you will find in this case .rad file is simply
0That means radial density is empty (no data), so there is no problem.
Offline
#32023-06-19 09:55:51
- amin.alibakhshi@hotmail.c
- Member
- Registered: 2019-08-25
- Posts: 23
Re: some potential bugs
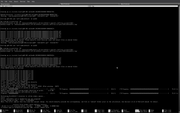
thank you for your quick follow-up. About g16, you were right and it was a bug in my own code. Nevertheless, for H+1, I still have a problem with computations that are automatically done by Multiwfn. I could manually fix it by writing H+1.rad with only one entry "0" and place in atmrad director as you suggested but for automatic usage of Multiwfn, I still get an error for every molecule in which the H atom is present as can be seen in this screenshot:
Last edited by amin.alibakhshi@hotmail.c (2023-06-19 09:56:32)
Offline
#42023-06-19 10:57:04
Re: some potential bugs
First, please make sure that you are using the latest version of Multiwfn. Multiwfn updates quite frequently.
Second, from your screenshot it seems that Multiwfn was not properly running in parallel mode, please try to use Multiwfn in interactive way (rather than submitting to server via script). If you have to run Multiwfn on server via script, I suggesting at least manually generating .rad files first and provide them in atmrad folder.
Offline
#52023-06-19 11:34:07
- amin.alibakhshi@hotmail.c
- Member
- Registered: 2019-08-25
- Posts: 23
Re: some potential bugs
I cannot make it interactively an have to use scripts because I am dealing with thousands of compounds. But I did some quick fix in my scripts via copying H+1.rad file from example/atmrad directory to the local atmrad directory and it fixed the problem and seems like a general remedy because for H+1 it makes no dependence on method/level of theory.
Thanks again for the fantastic job and kind regards,
Amin
Offline